Java TreeView uses two file formats to represent data, both of which are tab-delimited text. The third file format is an xml formatted file which holds settings information for persistence (see the section called “Persistence”).
Java TreeView File Formats
-
Generalized CDT File (.cdt and .pcl)
-
Tree File (.atr and .gtr)
-
XML Settings File (.jtv and global settings file, see the section called “Global Settings”)
The use of tab-delimitted text makes these files easy to edit in spreadsheet programs such as Excel, as well as manipulate with other programs.
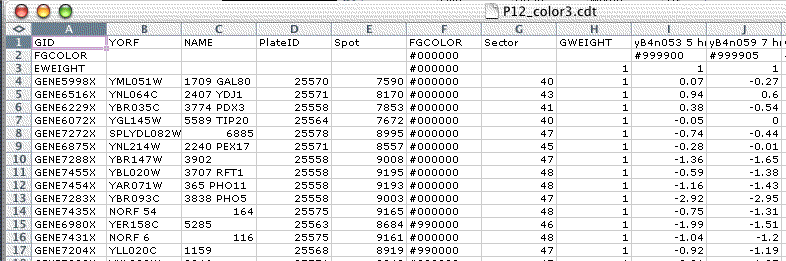
In order to view data in Java Treeview, there must be a Generalized CDT file. All other files are optional. The minimal GCDT file has a header row which contains the name of the unique id column, the name of the annotation column, and the names of the experiments, followed by one or more rows of per-gene data. Such a file can be created in Excel, and then saved as tab-delimited text.
The generalized CDT file is a straightforward generalization of the CDT and PCL file formats. In addition to expression data, this file can contain additional per-gene and per-array annotation in columns before the GWEIGHT column or in rows before the EWEIGHT row. For backwards compatibility, if the GWEIGHT column is missing Java TreeView assumes the data starts on the third column, or the fourth column if the first column has the header GID. Similarly, if the EWEIGHT row is missing Java TreeView assumes the data starts on the second row. As a general practice, it is a good idea to include the GWEIGHT column and EWEIGHT row.
In addition, Java TreeView does special things with the first two or three columns. If the first column is GID, the second and third are assumed to be the unique ID and NAME columns. If the first column is anything other than GID, the first and second columns are assume to be the unique ID and NAME columns. The unique ID is used for gene list export, and for some matching purposes when necessary. The NAME column is displayed as per-gene annotation in the dendrogram and other views.
There are annotation column names with special meaning to Java TreeView, and are used for coordinates or to set the color of gene names. These special columns are described after the basic file format, and should be avoided as annotation names unless you want that specific behavior.
A generalized CDT file is a tab-delimitted text file with the following specifications. The leftmost column and topmost row are reserved for headers. The file must contain at least two columns followed by a column with the header GWEIGHT, and at least one row followed by a row with the header EWEIGHT. Any rows and columns before the EWEIGHT and GWEIGHT are treated as annotation, and any after are treated as data. If a data value is missing or cannot be converted into a number, it is treated as not found. The annotation is kept in string form, and parsed by views as appropriate.
Some headers have special meaning to particular displays. This is a catalog of headers and their meaning to particular displays.
Table 2.1. CDT Column Headers With Special Meaning
| Header | Display | Meaning |
|---|---|---|
| FGCOLOR | Dendrogram | Color in which to render text for particular gene |
| BGCOLOR | Dendrogram | Color in which to render background for particular gene |
| LEAF | Dendrogram | The TIME at which this branch should be terminated. Used to indicate apopotosis in cell lineages, as well as phyogenetic distance in sequence alignments. See also TIME header for tree files. |
| CHROMOSOME | Karyoscope | The chromosome on which the gene is located, a natural number |
| ARM | Karyoscope | The arm of the chromosome, either "L", "R", or "1" meaning left, "2" meaning right. |
| POSITION | Karyoscope | The distance of the spot from the centromere in arbitrary units |
| GROUP | Dendrogram | Defines a partitioning of genes. The current (1.0.13) implementation of Dendrogram will insert a gap every time the GROUP value changes. At some point in the future, I may make it so that all genes with identical GROUP values are put in one cluster. |
Table 2.2. CDT Row Headers With Special Meaning
| Header | Display | Meaning |
|---|---|---|
| FGCOLOR | Dendrogram | Color in which to render text of array name |
| BGCOLOR | Dendrogram | Color in which to render background of array name |
| GROUP | Dendrogram | Defines a partitioning of arrays. The current (1.0.13) implementation of Dendrogram will insert a gap every time the GROUP value changes. At some point in the future, I may make it so that all rows with identical GROUP values are put in one cluster. |
In order for Karyoscope to correctly display gene expression data by chromosome location, it needs to know where exactly to position each unique ID. To this end, it looks for annotation columns with the names "CHROMOSOME", "ARM" and "POSITION", which designate the chromosome, arm and position of a particular gene. "CHROMOSOME" should be a natural number indicating which chromosome the unique ID is on, "ARM" should be either "R" or "L" indicating the arm, and "POSITION" should be a real number indicating how far from the centromere the unique ID is. There is really no restriction on the units for position; bp or kb are natural choices.
A coordinates file is simply a generalized CDT file which has such columns. The coordinates files supplied with Java TreeView do not contain any expression data; they consist entirely of the unique id column, the chromosome, arm and position columns, and the required GWEIGHT column. However, any other generalized CDT file with the correct columns can serve as a coordinates file.
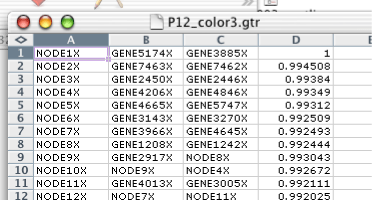
Traditionally, tree files have no header, and consist of four columns. Each row represents a node in either a gene tree, for the GTR file, or an array tree in the ATR file. For each row, the first column is the identifier of the node, the second column is the left child of the node, the third column is the right child, and the fourth column is the correlation between the left and right child. This fourth column is used by Java TreeView to determine the height of the node when rendering a tree.
By analogy to the CDT file, the tree files have been generalized in Java Treeview. Generalized tree files have a header line identifying the different columns. All generalized GTR/ATR files must have NODEID as the name of the first column. Tree files with any other string in the first row of the first column will be treated as legacy tree files. All of the rows will be treated as defining nodes, and the headers will be assigned the headers "NODEID", "LEFT", "RIGHT" and "CORRELATION". The meaning of these headers, and others, are described in the section called “ Tree File Headers ”.
All tree file headers can be displayed as node annotation in the dendrogram view. After loading a file in java treeview, select "Settings->Annotations..." to open the annotations dialog. Click "Gene Nodes" or "Array Nodes" and then select the desired headers. If you then mouse over the tree, the annotation for the select node will appear in the info pane.
Headers that have meaning beyond being used as annotation are described here.
Table 2.3. GTR/ATR Column Headers With Special Meaning
| Header | Display | Meaning |
|---|---|---|
| NODEID | Dendrogram | The value in this column serves as the identifier for the node. It must be unique. |
| LEFT | Dendrogram | The NODEID of the left child of this node. If the left child is not an internal node but a gene from the CDT file, the value should be the gene identifer, i.e. the value in the first column of the CDT file. |
| RIGHT | Dendrogram | The NODEID of the left child of this node, with leaf nodes handled as for the LEFT column. |
| CORRELATION | Dendrogram | The correlation value for this node. If this column appears, the implication is that the nodes in the tree should be arranged with a value of -1 most distal from the data matrix, with a value of 1 immediately proximal. This column and the TIME column should not appear in the same tree file. |
| TIME | Dendrogram | The time at which this node branched. Java Treeview is used by some to display cell lineages. For this purpose, it is useful to arrange branches in the tree by time at which the cell divided. This column is also used for indicating branching in sequence alignments, although TIME means phyogenetic divergence time in that context. If this column appears, the implication is that the nodes in the tree should be arrange with a value of 0 most distal from the data matrix, and that the node with maximum time value should be immediately adjacent to the data. |
| NODECOLOR | Dendrogram | The color value for this node. In the abscence of this column, the dendrogram is rendered black when not selected. Even when this column appears, selected nodes are rendered red. |